Are we entering a new era for clinical data analysis? An era in which massive time savings, quality improvements, and cost savings can be achieved by automation of the production of tables, listings, and figures (TFLs) in clinical trial analytics results. The holy grail of every Stats Programming team, surely!
The fast-evolving field of generative AI (GenAI) is capable of amazing feats of code generation. Inform those models with structured digital documents (not digitised documents) to define the target outputs; and inform those models with guidelines and constraints provided by standards, metadata, and models. The result will be high-quality outputs, simply requiring QA by a human (or perhaps an independent AI model?), for at least 80% of planned outputs.
“How could I get started?”, I hear you ask, well, let’s consider the three aspects of this trinity in turn.
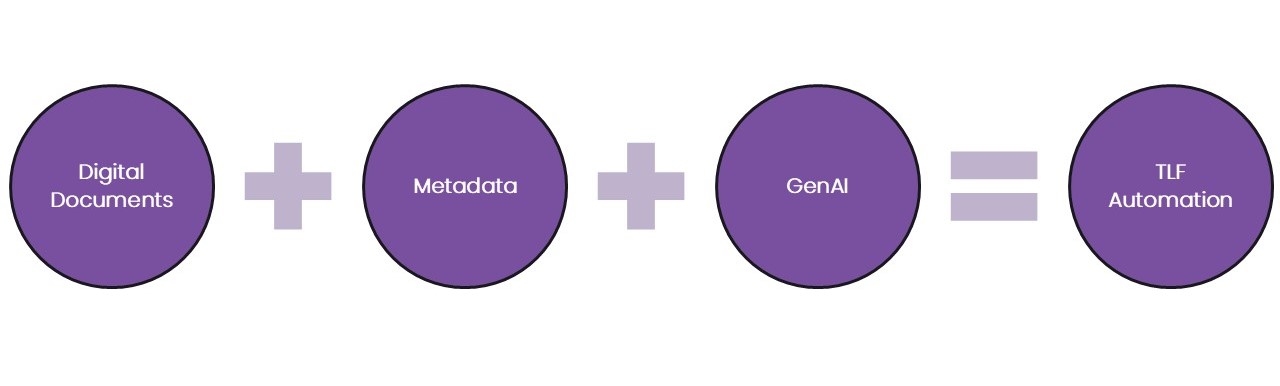
Digital Clinical Trial Documents
The first pillar in our trinity is digital documents, meaning our specifications.
The Transcelerate organisation is driving collaboration across biopharma R&D to identify ways to accelerate the delivery of new medicines. One aspect of this is to harmonise and digitise study documents via the Clinical Content and Re-use (CC&R) initiative.
This ongoing initiative has created a set of templates for Protocol, SAP, and Clinical Study Reports that allow for the creation of electronic, machine-readable documents. This opens up the possibility of reusing content in downstream processes, such as TLF generation.
The SAP, or Statistical Analysis Plan, is the key document here. It details how clinical data analysis will be performed – methods, population size, endpoints, etc., which drives the generation of ADaM datasets. It also details the Tables, Figures and Listings to be created, hopefully with associated table shells to give our programmer a lead on what they should produce; or maybe the GenAI can generate those mock shells too
Having a machine-readable SAP which is consistent across studies will improve the chances of our GenAI creating tables, listings and figures that we can use.
Metadata
Metadata – data that describes data, is our second pillar. It’s crucial for informing any programmer, human or computer, about the structure of the data they are working with.
We all know about CDISC and ADaM standards. We strive to develop our clinical data analysis datasets in compliance with CDISC standards, and indeed, datasets are required to be compliant for submission to the FDA. However, the flexibility within the ADaM standards means that datasets can be constructed differently between studies, yet still be technically compliant!
Having your ADaM data be compliant with CDISC standards and, more importantly, consistent across studies, at least within each Therapeutic Area (TA), will again improve their usability by your GenAI model and thus improve the quality of the programs produced.
You could, and should also use your existing programs, macros, and outputs as training data for your GenAI. We’ll learn more about this in the next section of this paper.
GenAI
The final, and perhaps most important pillar, is the GenAI model.
Our common experience of GenAI is likely to be something like ChatGPT or Co-Pilot. The author’s past attempts to ask ChatGPT to create snippets of R code have not been wholly successful when it came to execution. Which doesn’t sound very promising. However, it’s possible to train a GenAI model using our existing repository of programs, macros and outputs.
‘Public’ GenAI models would surely require us to share our code and data for training, but we all appreciate the sensitivity to clinical trial data sharing within our industry. Thankfully, it’s possible to implement your own internal GenAI model within your company network, trained on internal company materials plus a selection of external (public) materials.
Before you build your internal GenAI however, there are a few factors to consider:
- Which model
- Language choice
- Training the GenAI
- Regulatory Agency Guidance
Model Choice
Not all GenAI models are the same. Models exist that are already trained on coding languages including python, R and SAS. Be aware though that these models are trained on publicly available code repositories like Github where SAS is less common. So, while the model may be able to produce SAS code it will almost certainly require training on your own data to make output of better quality.
Examples of GenAI models include
GPT3 – a large language model (LLM) that can generate text and code, including SAS code.
CodeBERT – a pre-trained language model specifically designed to understand and generate code.
Language
As mentioned above, you will need to consider which language you want GenAI to use when creating your code.
R and Python have large user bases, and by nature of them being open source there is more focus on knowledge sharing. Thus, GenAI models have a wider knowledge base on which to draw.
SAS in contrast has a smaller user community and the complexities of the code syntax can make it harder for GenAI to create correctly functioning code first time. On the other hand, you’ve probably invested in a library of SAS macros which simplify the coding and produce corporate-standard outputs.
Training
Even a model that is already developed for coding will need to be trained to
- recognise the SAP and metadata as the inputs for the generation of TLFs
- learn the coding methods previously used by programmers to manipulate data and generate output
- understand how your standard macros work
- learn the desired layout and format of outputs
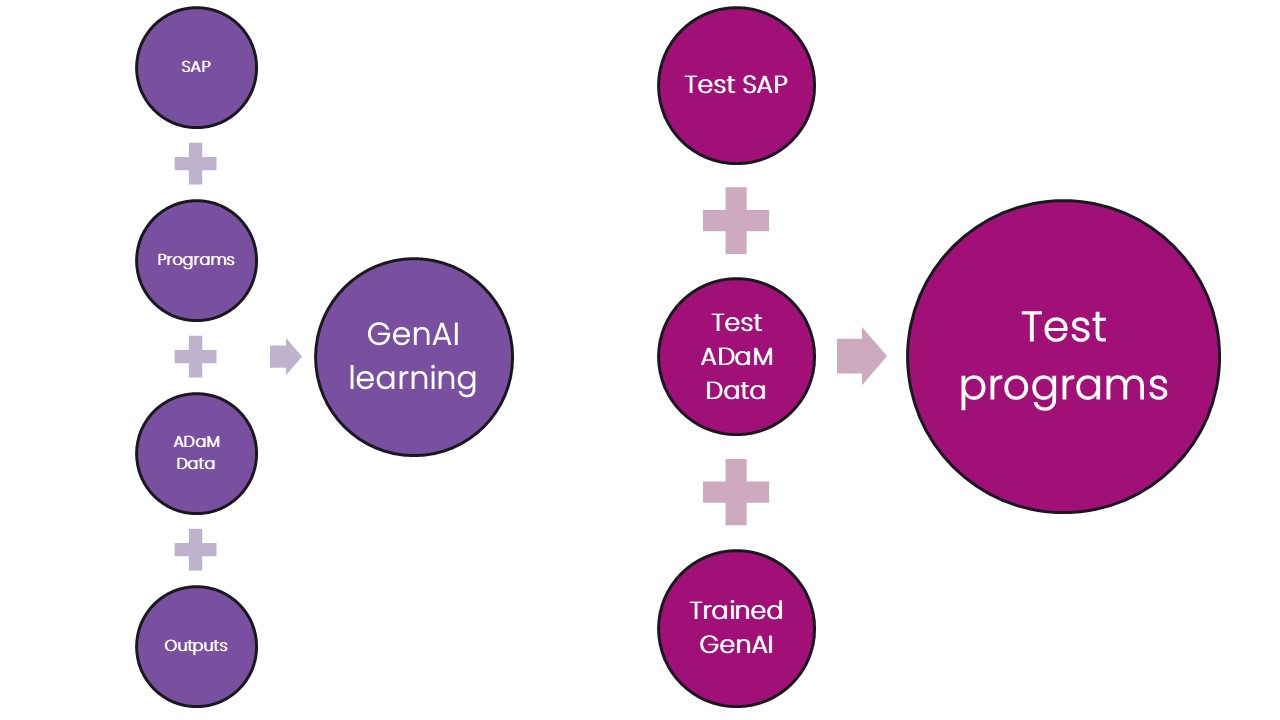
Divide your training data into two parts: training and testing sets. The training set allows the GenAI model to learn the patterns and relationships between the inputs, programs, and outputs. The larger your training set the better.
The testing set can then be used to test the newly trained model. Given a set of inputs – SAP table shells and ADaM datasets, can it produce code to create outputs that match the real thing? Note that if the performance in not acceptable then a different testing set should be used for a subsequent test.
Training your model should be a continuous process – as more studies are successfully reported the associated metadata should be fed into your model to improve its performance.
Regulatory Guidance
The EMA[1] and FDA[2] have both published papers on the use of Artificial Intelligence in the development of medicinal products. Neither document should be considered as regulatory guidance or policy, but they both contain useful information on what the regulatory agencies are thinking with regard to the use of AI.
They remind us that it is the responsibility of the clinical trial sponsor to ensure that all algorithms, models, datasets, and data processing pipelines used are fit for purpose and are in line with legal, ethical, technical, scientific, and regulatory standards as described in legislation, GxP standards and current regulatory guidelines. And note that these guidelines may be stricter than what is considered to be standard practice within data science.
When considering the implementation of GenAI within your organisation you should conduct a regulatory impact and risk assessment of all AI applications and contact regulatory agencies (e.g. the FDA CDER AI Steering Committee (AISC) where no clear written guidance exists.
It is recommended to read both papers as they provide useful information and references beyond the scope of this paper.
Conclusion
GenAI technology certainly offers an exciting opportunity to automate some TLF generation. Publicly available models won’t have been trained on clinical trial data so it’s recommended to implement a model within your company network, which will also alleviate concerns about making proprietary data available to the wider world.
However, GenAI is not infallible and so even after thorough training of your model, the code generated should be independently tested for accuracy.
And finally, remember to take into account the advice from the regulatory agencies.
Reach out and talk to an expert if you have any questions about clinical trial analytics, engaging with regulatory agencies, or anything else mentioned in this blog!
[1] https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-use-artificial-intelligence-ai-medicinal-product-lifecycle_en.pdf
[2] Using Artificial Intelligence & Machine Learning in the Development of Drug and Biological Products


