This blog explores how protocol-driven (Q)SAR assessments became routine in ICH M7-based impurity evaluations.
How ICH M7 Is Applied: Practical Steps for (Q)SAR Mutagenicity Assessment
It has been 9 years since the publication of the Principles and Procedures for implementation of ICH M7 recommended (Q)SAR analyses1, a paper Leadscope (Instem) helped initiate and promote. This publication has played an important role in shaping how drug impurities are assessed using in silico approaches. The publication, developed through cross-industry collaboration, provided practical guidance on implementing (Q)SAR predictions, incorporating expert review, and documenting assessments in alignment with the ICH M7 guideline2.
This initial publication paved the way for another pivotal paper3, focused on supporting indeterminate or out-of-domain results, which are two challenges frequently encountered in regulatory assessments. Together, these papers laid the foundation for a standardized approach that has been widely adopted across industry and regulatory workflows. Notably, both papers are open access and have been cited extensively, including in the ICH M7 Q&A document4, which highlights their continued relevance and utility.
Tools Supporting ICH M7 Class Assessments
The impact of these publications has extended beyond written publications. The principles and workflows have been implemented in software tools5 that support both internal industry assessments and regulatory submissions. For example, Leadscope’s ICH M7 protocol implementation integrates multiple prediction systems (statistical QSAR models, expert alerts) and database sources into a unified, documented decision framework to derive the M7 class assignment, Figure 16. This implementation streamlines the assessment process while supporting expert review with traceability.
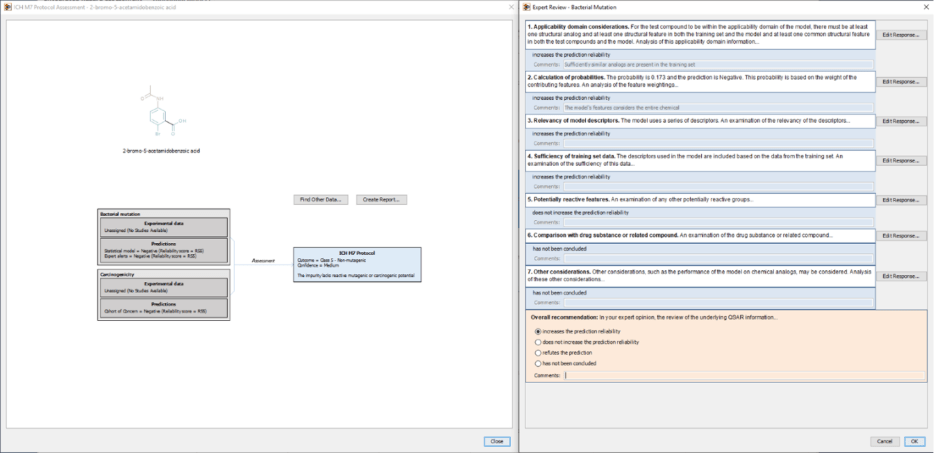
Figure 1: ICH M7 interactive decision scheme summarizing QSAR results and expert review framework
Documented Procedures for Other Endpoints
Importantly, the ICH M7 protocol implementation builds on the broader framework for in silico toxicology protocols developed and published in recent years. This includes similar structured workflows for endpoints such as genetic toxicology7 and skin sensitization8, and endocrine activity9. Each protocol follows a consistent architecture that enables transparency, repeatability, and integration of both model outputs and expert input.
ICH M7 continues to influence other regulatory sectors as well, with similar approaches being applied to the assessment of genotoxic or mutagenic impurities in animal health products10 , E&Ls, and pesticide residues11. Additionally, the foundation laid by the original principles and procedures publications1,3 has enabled these applications by demonstrating how protocol-driven approaches can be both scientifically rigorous and practical. The development of protocol-driven tools has played a critical role in ensuring that assessments are not only scientifically robust but also consistent, efficient, and defensible, which are important in a regulatory environment.
Key Takeways:
- The ICH M72 guideline has driven the adoption of standardized methodologies supporting (Q)SAR assessments.
- Standardized methodologies1,3 have been incorporated into (Q)SAR protocols to support impurity assessments (ICH M7) and additional use cases in a scientifically rigorous manner.
- Leadscope’s ICH M7 solution adheres to the established principles and procedures outlined in ICH M7 for impurity analysis.
You can access the seminal publications and the standardized methodologies for implementing (Q)SAR assessments within ICH M7 framework here:
Principles and procedures for implementation of ICH M7 recommended (Q)SAR analyses.
References:
2. ICH, 2017. ICH guideline M7 (R1). Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk (No. EMA/CHMP/ICH/83812/2013), ICH Harmonised Guideline. European Medicines Agency.
https://database.ich.org/sites/default/files/M7_R1_Guideline.pdf
3. A. Amberg et al., 2019, Principles and procedures for handling out-of-domain and indeterminate results as part of ICH M7 recommended (Q)SAR analyses, Regul. Toxicol. Pharmacol. 102, 53–64. https://doi.org/10.1016/j.yrtph.2018.12.007.
4. ICH M7 Guideline: Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk. Questions and Answers. https://www.ich.org/page/multidisciplinary-guidelines#7-3
5. https://www.instem.com/solutions/in-silico/leadscope-model-applier/
6. Myatt, G. J., Bassan, A., Bower, D., Johnson, C., Miller, S., Pavan, M., & Cross, K. P. (2022). Implementation of in silico toxicology protocols within a visual and interactive hazard assessment platform. Computational Toxicology, 21, 100201. https://doi.org/https://doi.org/10.1016/j.comtox.2021.100201
7. Hasselgren, C., Ahlberg, E., Akahori, Y., Amberg, A., Anger, L. T. L. T., Atienzar, F., Auerbach, S., Beilke, L., Bellion, P., Benigni, R., Bercu, J., Booth, E. D. E. D., Bower, D., Brigo, A., Cammerer, Z., Cronin, M. T. D. M. T. D., Crooks, I., Cross, K. P. K. P., Custer, L., … Myatt, G. J. G. J. (2019). Genetic toxicology in silico protocol. Regulatory Toxicology and Pharmacology, 107. https://doi.org/10.1016/j.yrtph.2019.104403
8. Johnson, C., Ahlberg, E., Anger, L. T., Beilke, L., Benigni, R., Bercu, J., Bobst, S., Bower, D., Brigo, A., Campbell, S., Cronin, M. T. D., Crooks, I., Cross, K. P., Doktorova, T., Exner, T., Faulkner, D., Fearon, I. M., Fehr, M., Gad, S. C., … Myatt, G. J. (2020). Skin sensitization in silico protocol. Regulatory Toxicology and Pharmacology, 116, 104688. https://doi.org/https://doi.org/10.1016/j.yrtph.2020.104688
9. Johnson, C., Marty, S., Kim, M., Crofton, K., Roncaglioni, A., Bassan, A., Barton-Maclaren, T., Domingues, A., Frericks, M., Karmaus, A., Kulkarni, S., Piparo, E. lo, Melching-Kollmuss, S., Tice, R., Woolley, D., & Cross, K. (2025). An in silico protocol for endocrine activity assessment: Integrating predictions, experimental evidence, and expert reviews across estrogen, androgen, thyroid, and steroidogenesis modalities. Computational Toxicology, 100364. https://doi.org/https://doi.org/10.1016/j.comtox.2025.100364
Please contact us to speak to an expert or learn more about our solutions here.


